The problem
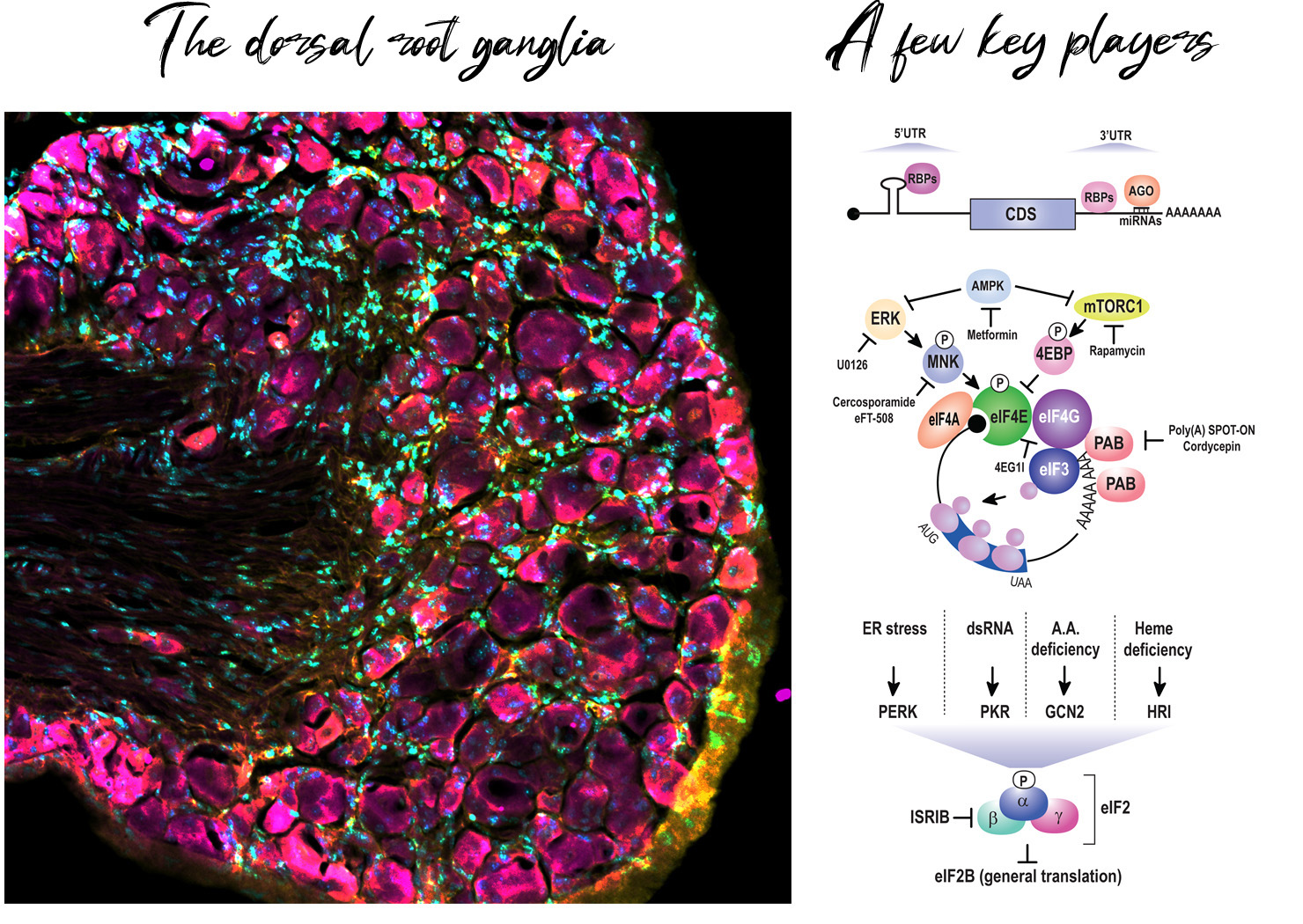
Pain is pervasive and devastating. Poorly treated chronic pain is the largest source of disability in America with an estimated economic cost of more than 500 billion dollars per year. Moreover, existing strategies to disrupt pain (e.g. opiates) have well known and highly undesirable side effects. Better strategies for preventing chronic pain are desperately needed.
In the vast majority of cases, pain originates in the periphery in a specialized type of sensory neuron called a nociceptor. Long-lived changes in their excitability – which are intimately liked to chronic pain – require de novo protein synthesis. Multiple groups have shown that inhibition of translation diminishes pain associated behaviors in relevant pre-clinical models.
Our overaching goal is to understand how mRNAs are controlled in nociceptors. To achieve this, we probe fundamental molecular mechanisms that govern pain-associated plasticity in pre-clinical rodent and human iPSC models. This work has already resulted in the identification of new targets. Several lines of investigation we are pursuing are described below.
Translation
We are interested in understanding how translation is controlled in sensory neurons. Relevant publications:
1. Smith PR, Garcia G, Meyer AR, Ryazanov AG, Ma T, Loerch S, Campbell ZT. eEF2K regulates pain through translational control of BDNF. Molecular Cell. 2025. In press. PMID: 39694034
2. de la Peña JB, Kunder N, Lou TZ, Chase R, Stanowick S, Barragan-Iglesias P, Pancrazio JJ, Campbell ZT. A role for translational regulation by S6 kinase and a downstream target in inflammatory pain. British Journal of Pharmacology. 2021 Dec;178(23):4675-4690. PMC9169231
3. Smith PR, Loerch S, Kunder N, Stanowick AD, Lou TF, Campbell ZT. Functionally distinct roles for eEF2K in the control of ribosome availability and p-body abundance. Nature Communications. 2021. 12(1):1-16. PMC8611098
For an detailed explaination of translational control in pain, please click here
Functional genomics
We leverage a range of sequencing approaches to understand gene control in the sensory neurons. Relevant publications:
1. Chase R, de la Peña JB, Smith PR, Lawson J, Lou TF, Stanowick AD, Black BJ, Campbell ZT.Global analyses of mRNA expression in human sensory neurons reveal eIF5A as a conserved target for inflammatory pain. FASEB J. 2022 Jul;36(7):e22422. PMID: 35747924
2. de la Peña JB, Barragan-Iglesias P, Lou TF, Loerch S, Kunder N, Shukla T, Song J, Megat S, Moy JK, Wanghzou A, Ray PR, Hoyt K, Steward O, Price TJ, Shepherd J, Campbell ZT. Intercellular Arc signaling regulates vasodilation. J Neuroscience. 2021. 15;41(37):7712-7726. PMID: 34326146
3. Barragan-Iglesias P, Kunder N, Wanghzou A, Black B, Ray PR, Lou TF, de la Peña JB, Atmaramani R, Shukla T, Pancrazio JJ, Price TJ, Campbell ZT. A peptide encoded within a 5' untranslated region promotes pain sensitization in mice. Pain. 2021. 162(6):1864-1875. PMC8119312
mRNA stability
We are interested in probing how mRNA stability, storage, and localization contribute to pain signaling. Relevant publications:
1. Iglesias P, Lou TF, Bhat V, Megat S, Burton M, Price T, Campbell ZT. Inhibition of Poly(A)- Binding Protein with an RNA mimic reduces pain sensitization in mice. Nature Communications. 2018. 9(1):10. PMC5750225
2. June Bryan de la Peña, Rebecca Chase, Nikesh Kunder, Patrick R Smith, Tzu-Fang Lou, Alexander Stanowick, Prarthana Suresh, Tarjani Shukla, Samuel E Butcher, Theodore Price, Zachary T Campbell. Inhibition of nonsense-mediated decay induces nociceptive sensitization through activation of the integrated stress response. J Neuroscience. 2022. PMID: 36894318
3. Kunder N, de la Peña JB, Lou TF, Chase R, Suresh P, Lawson J, Shukla T, Black BJ, Campbell ZT. The RNA-binding protein HuR is integral to the function of nociceptors in mice and humans. J Neuroscience. 2022. 42(49):9129-9141. PMID36270801
RNA-protein interactions
Recognition of RNA by proteins is the prerequisite for all subsequent regulation and is critical to decipher. Relevant publications:
1. Campbell ZT, Valley CT, Wickens M. A protein-RNA specificity code enables targeted activation of an endogenous human transcript. Nature Struct Mol Biol. 2014. (21): 732-738. PMC4125476
2. Zhou Q, Kunder N, De la Paz JA, Lasley AE, Morcos F, and Campbell ZT. Global pairwise RNA interaction landscapes reveal core features of protein recognition. Nature Communications. 2018. 9(1):2511. PMC6023938
3. Bhat VD, McCann KL, Wang Y, Fonseca DR, Shukla T, Alexander J, Qiu C, Wickens M, Lo TW, Hall TT, Campbell ZT. Engineering a conserved RNA regulatory protein repurposes its biological function in vivo. eLife. 2019. 8 e43788. PMC6351103
An up to date list of published work can be found here
Papers
The most up to date list of our papers can be found on Pubmed or on our Google scholar page
Selected references:
1. Smith PR, Garcia G, Meyer AR, Ryazanov AG, Ma T, Loerch S, Campbell ZT. eEF2K regulates pain through translational control of BDNF. Molecular Cell. 2025. In press. PMID: 39694034
2. Smith PR, Loerch S, Kunder N, Stanowick AD, Lou TF, Campbell ZT. Functionally distinct roles for eEF2K in the control of ribosome availability and p-body abundance. Nature Communications. 2021. 12(1): 1-16. PMC8611098
3. Iglesias P, Lou TF, Bhat V, Megat S, Burton M, Price T, Campbell ZT. Inhibition of Poly(A)- Binding Protein with an RNA mimic reduces pain sensitization in mice. Nature Communications 2018. 9(1):10. PMC5750225
4. Kunder N, de la Peña JB, Lou TF, Chase R, Suresh P, Lawson J, Shukla T, Black BJ, Campbell ZT. The RNA-binding protein HuR is integral to the function of nociceptors in mice and humans. J Neuroscience. 2022. In press. PMID36270801
Open positions
We are currently recruiting postdocs and graduate students from multiple programs including: MCP, NTP, iPIB, and CMB. To apply, please email the PI a copy of your CV.
Current lab members

Zachary Campbell, Bamforth Distinguished Chair in Anesthesiology, Vice Chair for Research, and Associate Professor Department of Anesthesiology University of Wisconsin School of Medicine and Public Health, Lab head

Guadalupe Garcia Perez, PhD CINVESTAV-IPN, Postdoctoral fellow

Alex Mikesell, PhD Medical College of Wisconsin, Postdoctoral fellow

Angela Meyer, BS Purdue, iPIB graduate student

Jake Shapiro, BS Colorado College, iPIB graduate student

Sydney O'Flanagan, Genetics undergraduate student, research intern

Aleena Varghese, Molecular and Cellular Biology undergraduate student, research intern
Alumni
*This information reflects known positions after training and may not be current
Former postdocs
Prof. June Bryan De La Pena - Assistant Professor UNM, Department of Anesthesiology
Prof. Bryan Black (co-mentored with Dr. Joseph Pancrazio) - Assistant Professor UMass-Lowell, Department of Biomedical Engeneering
Prof. Paulino Barregan Iglesias (co-mentored with Dr. Price) - Assistant Professor Universidad Autónoma de Aguascalientes
Dr. Chi Zhang - Assistant Instructor, UT-Southwestern
Former doctoral students
Dr. Pat Smith (PhD 2024) - Postdoc Dr. Amy Lee, Harvard
Dr. Rebecca Chase (PhD 2023) - Postdoc Dr. Joe Dougherty, WashU
Dr. Nikesh Kunder (PhD 2022)- Postdoc Dr. Jason M. Schenkel, MD Anderson
Dr. Vandita Bhat (PhD 2020) - Postdoc Dr. Guang Hu, NIEHS Stem Cell Biology Group
Former masters students
Prathana Sursh (MS 2022) - PhD student, UT-Dallas
Sanjana Rajeev (MS 2018) - PhD student, USC
Alexa Lasley (MS 2018) - scientist at nanostring
Former staff scientists
Tzu-Fang Lou - Core facility director, UT-Dallas
Jane Song - PhD student, CU-Boulder
Eric Wistrom - PhD student, teaching abroad in Provence
Sam Rovetto - Scientist at Labcorp
Former undergraduates
Lexi Sharkus - RUSCH research intern, summer 2023, applying for graduate programs
Varun Kotipalli - undergraduate student at UT-Dallas
Amber Ho - Pharmacovigilance Specialist
Alex Stanowick - graduate student at UT-Dallas
David Kim - Applying to graduate programs
Krishnateja Kommera - medical student
Siddhardha Maligireddy - postbac NIH intramural researcher NCI
Miloni Joshi - Medical scribe
Emma Henderson - Biomedical engineer at ICA
Ali Abbas - bussiness analyst
Anaid Cándido - Medical Education Quality Officer at National health service UK
Cori Weaver - Operations Manager at Metis Genetics
Dallas Fonseca - Graduate student student University of Minnesota
Sulaiman Syed - Medical Student at University of the Incarnate Word School of Osteopathic Medicine
Abril Ruiz - Clinical Research Associate at Icon
Andrea Kibel - Graduate student University of Notre Dame
Prerana Ramadurgum - DO student UNT Health Science Center
Sanjana Rao - Senior Associate Software Engineer
Neil Mascarenhas - Student at Texas A&M College of Dentistry
Tejesh Guddanti - Medical Student at University of Oklahoma
Contact
You can reach the lab head at this email address: zcampbell /at/ wisc /dot/ edu
Elements
Text
This is bold and this is strong. This is italic and this is emphasized.
This is superscript text and this is subscript text.
This is underlined and this is code: for (;;) { ... }. Finally, this is a link.
Heading Level 2
Heading Level 3
Heading Level 4
Heading Level 5
Heading Level 6
Blockquote
Fringilla nisl. Donec accumsan interdum nisi, quis tincidunt felis sagittis eget tempus euismod. Vestibulum ante ipsum primis in faucibus vestibulum. Blandit adipiscing eu felis iaculis volutpat ac adipiscing accumsan faucibus. Vestibulum ante ipsum primis in faucibus lorem ipsum dolor sit amet nullam adipiscing eu felis.
Preformatted
i = 0;
while (!deck.isInOrder()) {
print 'Iteration ' + i;
deck.shuffle();
i++;
}
print 'It took ' + i + ' iterations to sort the deck.';
Lists
Unordered
- Dolor pulvinar etiam.
- Sagittis adipiscing.
- Felis enim feugiat.
Alternate
- Dolor pulvinar etiam.
- Sagittis adipiscing.
- Felis enim feugiat.
Ordered
- Dolor pulvinar etiam.
- Etiam vel felis viverra.
- Felis enim feugiat.
- Dolor pulvinar etiam.
- Etiam vel felis lorem.
- Felis enim et feugiat.
Icons
Actions
Table
Default
| Name |
Description |
Price |
| Item One |
Ante turpis integer aliquet porttitor. |
29.99 |
| Item Two |
Vis ac commodo adipiscing arcu aliquet. |
19.99 |
| Item Three |
Morbi faucibus arcu accumsan lorem. |
29.99 |
| Item Four |
Vitae integer tempus condimentum. |
19.99 |
| Item Five |
Ante turpis integer aliquet porttitor. |
29.99 |
|
100.00 |
Alternate
| Name |
Description |
Price |
| Item One |
Ante turpis integer aliquet porttitor. |
29.99 |
| Item Two |
Vis ac commodo adipiscing arcu aliquet. |
19.99 |
| Item Three |
Morbi faucibus arcu accumsan lorem. |
29.99 |
| Item Four |
Vitae integer tempus condimentum. |
19.99 |
| Item Five |
Ante turpis integer aliquet porttitor. |
29.99 |
|
100.00 |








